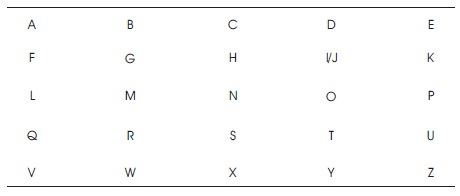
Table 1. Key Matrix in Traditional Playfair
DNA cryptography is one of the cryptographic techniques in which DNA is used to store and transmit the information. With the strange arrangement of information in DNA sequences, it is possible to implement data security efficiently. Though a lot of research is done and many algorithms have been developed for hiding the data, DNA sequences based data encryption seems to be an efficient approach for satisfying the present information security needs. In this paper, the traditional Playfair cipher to DNA Cryptography is applied. The key matrix of traditional matrix contains the 25 alphabets of the total 26-alphabets in a 5x5 key matrix based on the secret key. In the present paper, the key matrix uses 64 DNA codons by arranging them into 8x8 matrix so that the security is enhanced. The encryption technique has been modified, but at the same time retains some of the basic rules of the classical Playfair Algorithm. Playfair technique is modified by arranging the DNA Codons into 8x8 matrix to eliminate its limitations and enhance its security features. The proposed algorithm is efficient as it contains a LOOKUP table, which contains 64 values (A..Z,a..z,0..9, ,.) And the LOOKUP table is randomly arranged for each transmission. The proposed method is implemented and also compared with other popular ciphers on the basis of Time Complexity. The main objective of this paper is to provide a security system which uses DNA as a media.
We all know providing security to the information is a key aspect in the field of Computer Science. With the growth of technology, threats has also increased. Designing new algorithms for a secured transmission system is the need of the hour. Human beings created an innovation in which the transmitted data looks unreadable thereby the attacker does not mean what it is. But the attacker tries various mechanisms to break the algorithms. The main feature of cryptography is to have Confidentiality, Authentication, and Integrity of the data. The process of converting a plaintext into a cipher text is called encryption and vice versa is called decryption. The encryption and decryption takes place by taking one or more keys. Based on the key type, the security mechanism is used. During the process if only one key is used for both encryption and decryption process, then it is called symmetric key cryptography. Alternately, if two keys are used, one is for encrypting (Public Key) and the other is for decrypting (Private Key), then it is called as Asymmetric Key Cryptography. As many cryptographic algorithms are cracked, researchers are continuously striving to develop strong algorithms to provide security. Performing computation with DNA sequences is DNA Computing. Adleman (1994) and Gehani et al. (2003) have identified that DNA can be used as one of the data hiding techniques in the field of DNA cryptography. The idea behind this is introducing new methodologies to give high security along with conventional cryptography. Initially, the information is stored in DNA sequence and then computation is applied on the DNA sequence. The main advantage of DNA computers is to generate more feasible solutions simultaneously to solve the complex problems. It is known as Parallel Processing (Anam et al., 2010; Lipton, 1995). Silicon based computers solve the problems by using linear processing. To increase the processing speed and to solve the complex problems at the lesser time, researchers introduced bio-chips which means microarray technology. It is very useful to store and handle the biological information (Clelland et al., 1999 ; Lai et al., 2010).
Playfair cipher is a symmetric encryption algorithm, which takes a sequence two adjacent characters in the plaintext as single units and converts them into cipher text (William, 2003). In the first step, the key is processed i.e., remove all the repeated characters from the key and arrange the remaining key values in a predefined 5x5 alphabet matrix (Table 1). There are 26 alphabets in English and these alphabets can be arranged in 5x5 matrix and in one cell i and j characters are placed. Then process the plaintext, i.e., if it contains two consecutive letters then insert ‘X' in the middle. If the number of plaintext characters after inserting 'X' is odd then add 'X' in the last to make it even. Then split into two characters each and apply the following three rules for diagram.

Table 1. Key Matrix in Traditional Playfair
In Playfair algorithm, the key and plaintext restrict to 26 characters only and there is no possibility of taking any special character set. In order to overcome this, the authors have proposed DNA Playfair which is limited to 26 characters by considering the protein sequences.
DNA is Deoxyribonucleic acid which contains four nitrogen bases, five carbon sugar and phosphate group. It contains the genetic information of all living organisms. The four nitrogen bases Adenine (A), Thymine (T), Cytosine (C), and Guanine (G) can be used to encode the information which is transmitted in the network. According to the central dogma of molecular biology, the DNA is converted into RNA, called as transcription and the RNA is converted into Proteins, called as translation. In the transcription process each three DNA bases can be called as DNA Codon and is replaced with equivalent Amino Acid. But the reverse process cannot be done (Rakheja, 2011; Abbasy & Shanmugam, 2011). Due to this, many researchers used these codons to implement different cryptographic algorithms. Gao (2010) proposed biological alphabets to represent the information using DNA, RNA, and Protein. Ning (2009) described the encryption of plaintext using the concept of splicing, introns. The DNA data is translated into RNA which is converted into protein form of data and is sent to the receiver. Singh et al. (2010) have proposed DNA based cryptography to secure mobile networks by applying splicing mechanism. The algorithm is based on cutting of introns from the DNA sequence which is obtained from plain text. In next step the position is identified where the intron is cut. Then position and introns are combined to form a key file K1. So there is a conversion of DNA into mRNA and then a conversion of data into amino acid, which is to be transmitted to receiver. Selecting random key information from the DNA strand using splicing mechanism was proposed by Babu et al. (2016). A D-Box designed using codons and a cipher layer added to the traditional Feistel cipher gives better result and was proposed by Devi and Kumar (2018).
Sabry et al. (2010) have proposed a play fair cipher implementation using DNA Amino Acids. In this paper, the process is divided into two steps. The first step is Pre-processing, in which the secret key is prepared by removing spaces, repeated characters, and then the key is converted into uppercase. The plaintext is also pre-processed by removing all the spaces to avoid the attacker's trace. Then processing takes place in second step. Here, the plaintext is converted into binary and then it is converted into DNA bases. From the DNA sequence, consider the three bases as a codon and replace the codon into its equivalent amino acid. In protein synthesis, 20 amino acids are formed from 64 codons. As 26 alphabets exist in English language, these 20 amino acids are adjusted to these 26 alphabets. Further, ambiguity number is also noted to remove the ambiguity of the amino acids. Arrange the key value into 5x5 play fair matrix and apply the play fair cipher technique. The resultant code is DNA form of cipher text. Ambiguity is also converted into DNA base. The final cipher text is the concatenation of the DNA form of cipher text and ambiguity DNA base. In this algorithm, constructing the distribution of codon table is to get all the English alphabet letters and also keep track of the ambiguity value of each amino acid alphabet takes high time complexity (Table 2). The level of encryption process is high because it takes two levels to perform. It also had the drawback of storing the key size up to 25 characters only because the arrangement of key can be done in 5x5 matrices.
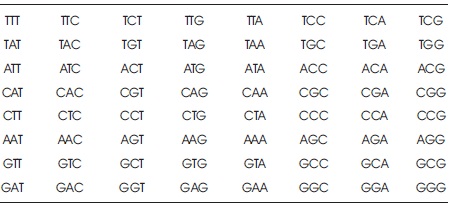
Table 2. Key Matrix of Proposed Algorithm
Atito et al. (2012) developed Playfair method using insertion technique by adopting the play fair and extended the work by implementing the algorithm in two levels. In the first level, the technique of Playfair was proposed by Sabry et al. (2010). In the next level, insertion technique is applied to the encrypted data of the first level. The cipher text is in the form of faked DNA sequence, but it contains hidden data. The authors have used 5x5 key matrix in this also.
To overcome this, the authors have proposed a new DNA based play fair cipher mechanism, which is based on the DNA LOOKUP table (Table 3) and also based on the arrangement of key matrix of size 8x8, which contains all the 64 codons. The LOOKUP table contains all the DNA Codons and each codon mapping with any character from the character set is defined as {A..Z, a..z, 0-9, space ,.}. In the proposed algorithm, the key is processed, and in processing stage, remove all the repeated characters from the key and map each character of the key into LOOKUP table value. After getting the DNA sequence, it is organised as a codon and arranged in 8x8 play fair matrix proposed in this algorithm. Now, process the plaintext. Add 'x' character to the plaintext if it contains two characters next to each other. Apply the rules of traditional play fair cipher.
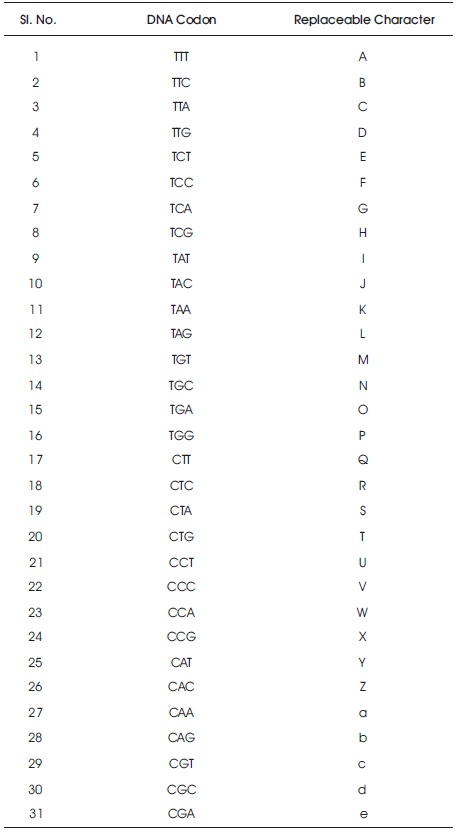
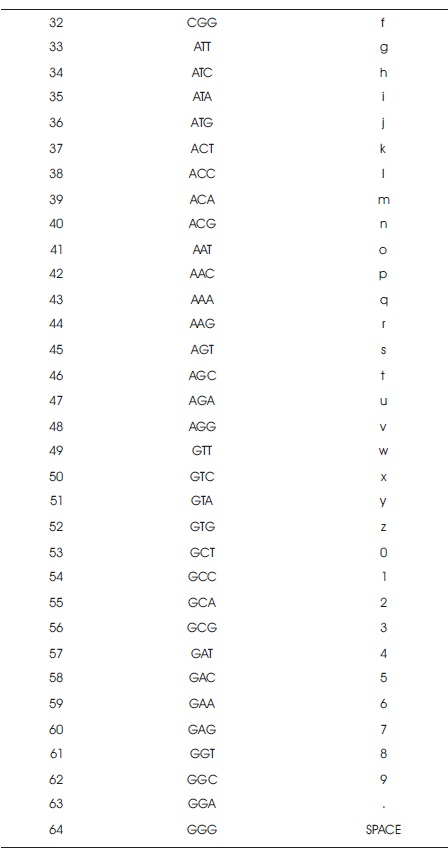
Table 3. LOOKUP Table
M is a plaintext and Key is the Secret Key which is in the set {A..Z, a..z, 0..9, ,.}*.
Begin
Step 1. Remove repeated characters from the key.
Step 2. The key can be converted into DNA bases from the LOOKUP Table (Table 3).
Step 3. Split the DNA bases into Codons.
Step 4. Construct a Playfair matrix of size 8x8 (Table 2) and the key codon values into row by row and add other codons.
Step 5. Remove the spaces and insert 'x' in the middle of the two consecutive same characters.
Step 6. Convert the plaintext into equivalent ASCII and in turn convert into DNA sequence.
Step 7. Split the DNA sequence into three bases each.
Step 8. Divide the Plaintext Codons pair wise.
Step 9. Apply Playfair algorithm to the above.
Step 10. Finally the Cipher text is sent to the receiver through public medium.
End
Take a plaintext M and the key K. Arrange LOOKUP table randomly with 64 codons and their equivalent character set {A..Z, a..z, 0..9, ,.). From the key, remove all repeated characters and match the equivalent codon from the character which is in the key. Now, construct a Playfair matrix of size 8x8 with the codon values row by row. In the plaintext, add 'x' in the middle of the two consecutive same characters and remove the spaces, if any. After that convert the plaintext into equivalent ASCII and then convert into DNA sequence. Divide the DNA sequences into Codons and then take the codons pairwise and apply the Playfair algorithm on those pairs. Finally, we get the ciphertext.
C is a Ciphet text and Key is the Secret Key.
Begin
Step 1. Remove repeated characters from the key.
Step 2. The key can be converted into DNA bases from the LOOKUP table (Table3).
Step 3. Split the DNA bases into Codons.
Step 4. Construct a play fair matrix of size 8x8 (Table 2) and Key codon values into row by row and add other codons.
Step 5. Divide the cipher text into codons.
Step 6. Apply Playfair Decryption algorithm to the above codons.
Step 7. Convert the DNA bases into equivalent binary and then convert them into equivalent ASCII character.
Step 8. Remove the 'x' which is placed in the middle of repeated characters.
End
The cipher text and the key are sent to the receiver. The receiver processes the key based on the LOOKUP table. Now, construct a Playfair matrix of size 8x8 with the codon values row by row. Divide the ciphertext into codons. Consider the codons into pairwise and apply the Playfair rules. After that DNA bases are converted into equivalent binary and in turn converts into equivalent ASCII character. The inserted 'x' values are removed from the plaintext.
Take an example of Plaintext=helloworld and the key=Playfair.
In the first step, process the key from the LOOKUP Table (Table 3). After removing the repeated characters from the key Playfair, we get playfair. Match these characters from the LOOKUP table (Table 3) and note down the codon values then the key is AAC, ACC, CAA, GTA, CGG, ATA, AAG and arrange these value in the key matrix (Table 2), then the output key matrix is shown in Figure 1.
In the second step, process the plaintext, i.e., if same characters are consecutive then insert 'x' in between the characters then the plaintext becomes helxloworld. Now check the length of plaintext. If it is odd add 'x' at the end of the string. In the current plaintext the length is 11 characters so add 'x' to the end of the string then the plaintext is helxloworldx.
In the next step, Convert the plaintext into ASCII, then into binary and then convert into DNA Digital Coding (00-A, 01- C, 10-G, 11-T).
Now, the DNA sequence is
CGGACGCCCGTACTGACGTACGTTCTCTCGTTCTAGCGTA CGCACTGA
Divide the sequence into codons then
CGG, ACG, CCC, GTA, CTG, ACG, TAC, GTT, CTC, TCG, TTC, TAG, CGT, ACG, CAC, TGA
Divide these codons pair-wise and apply Playfair encryption algorithm. In Playfair, consider the first two pairs CGG & ACG and find if these codons are placed in same row or same column or none of this in Key matrix of proposed algorithm after processing (Figure 2a). In the present case, the pair of codons are in same column then we take the codons beneath the actual position and then we obtain TCC and CTC.
Consider the next pair that is CCC & GTA and these are placed in different row and column values. Then we form the rectangle joining these codons as shown in Figure 2(b). The characters at the corners are replaced by the other pair of corners of the rectangle defined by the original pair as shown in Figure 2(c).
The codons at the corners are replaced by the other pair of corners of the rectangle defined by original pair as shown in Figure 2(d). Now, we got AAT & AAC. Do the same for all pairs.
The result sequence which is final cipher text is
C=TCCCTCAATAACCTCCACTGTAGGCTGTCCTTGTACATC CATCATTGG
The key matrix after processing and the final cipher text result is shown in Figure 2(e).
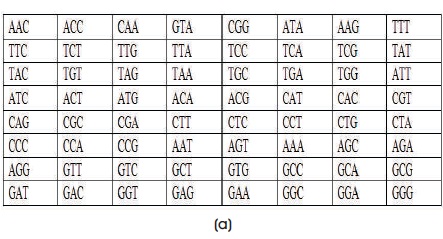
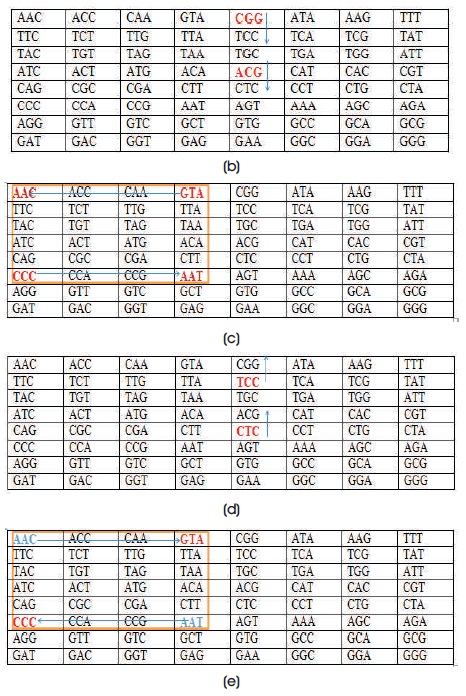
Figure 2. Key Matrix after Processing
The binary and ASCII value for digital coding is shown in Table 4.
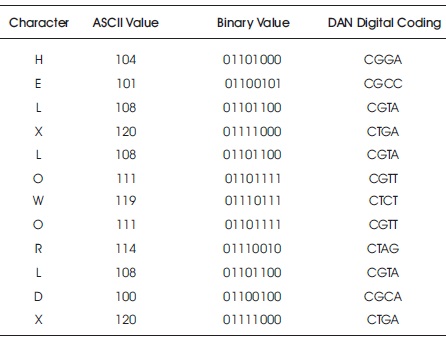
Table 4. ASCII and Binary Value for PAN
The receiver receives the key from the sender through secure channel. The procedure to process the key is same as in the encryption process.
The ciphertext at the receiver end is
C=TCCCTCAATAACCTCCACTGTAGGCTGTCCTTGTACATC CATCATTGG
Divide the cipher text into codons, i.e,
C=TCC,CTC,AAT,AAC,CTC,CAC,TGT,AGG,CTG,TCC,TTG,T AC,ATC,CAT,CAT,TGG
Now, divide the above codon sequence pairwise and apply decryption process of Playfair as shown in Figure 3. The first pair is TCC & CTC and check the positions of the codons into key matrix after processing (Figure 2a). If these two are in the same column, then we take the upper codons of their positions that is, CGG & ACG.
Consider the next pair, AAT & AAC, these pairs are not placed in same row and same column. Then, form a rectangle by joining these codons and take the corner codons and then we obtain CCC & GTA.
Do the same for the remaining codon pairs and we got final sequence as follows.
CGGACGCCCGTACTGACGTACGTTCTCTCGTTCTAGCGTA CGCACTGA
Convert the above sequence into binary sequence (A- 00, C-01, G-10, T-11)
01101000, 01100101, 01101100, 01111000, 01101100, 01101111, 01110111, 01101111, 01110010, 01101100, 01100100, 01111000
Convert the above sequence into ASCII which is further replaced by the equivalent character. The result is 104, 101, 108, 120, 108, 111, 119, 111, 114, 108, 100, 120 h, e, l, x, l, o, w, o, r, l, d, x.
Finally remove the padding character 'x' from the plain text to get the final text as helloworld.
The text has been taken as the input plaintext. The experiment is tested with various block sizes of 500, 1000, 1500, 2000, and so on characters. The key value is taken as “dnaplayfair”.
This method has been tested on NET framework 2008 on Intel core2Quad, 2.66 GHZ intelG31 motherboard, 320 GB HDD, 2 GB RAM. A comparison study is done by taking different lengths of character sets and the time complexities of encryption and decryption processes of proposed algorithm has been noted. Table 5 and Figure 4 shows better time complexity of the proposed method than previous methods. The time taken for loading LOOKUP table and processing the secret key is negligible.
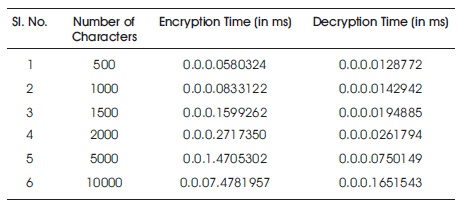
Table 5. Result Analysis
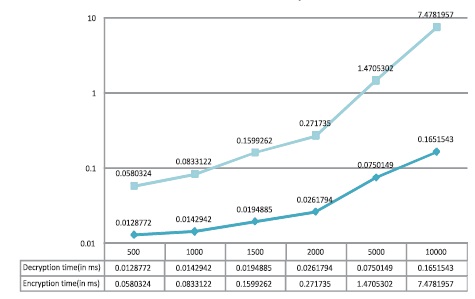
Figure 4. Performance Analysis of the DNA PlayfairCryptoSystem
DNA computers wipe out the traditional computers in future, as many scientists work to find the new material to perform the computation faster than the previous ones. One day DNA might be combined into computer chip to create bio-chip without harming the society. Not only this, DNA has the high capability to store huge amount information like terabytes in a single gram of DNA base. The proposed algorithm has encrypted the key based upon the LOOKUP table and the key not only contains alphabets, it contains uppercase, lower case alphabets, digits and special symbols like space and fullstop. Though the traditional play fair or DNA play fair is based on the 5x5 matrix and the key also limits to only alphabet characters, the proposed algorithm has taken the size of key matrix as 8x8. Based on the result analysis, this research proves to provide more security.