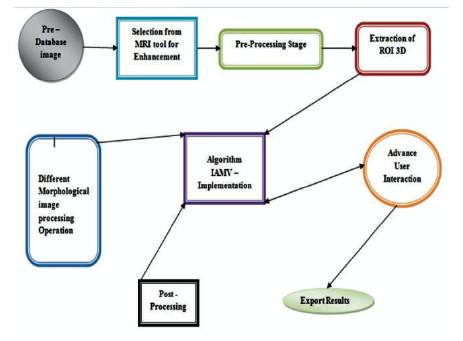
Figure 1 . Proposed Methodology Flow Diagram
The fusion expression means to extract a sequence which is acquired in several domains. The three-dimensional (3D) images have the deep information, which is not available in the conventional 2D images. The image fusion procedure of two images aim to get a more in-depth examination of the picture. 3D Fusion of medical images are found to be useful that they are medical images containing the data with significant scientific information for doctors during their analysis. The objective of this work is to examine the subsections of the obtained 3D structure along three axes. The paper deals with the DICOM (Digital Imaging and Communication in Medicine) images restoration, which is initially extremely useful for production of customized data that are atomically implemented by using a fast prototyping technology. MRI Images provide better contrast of soft tissues than CT images. Hence it provides better results in image fusion of MRI and CT images is done by using Wavelet Transforms in MATLAB. The researchers are forever focusing on biomedical 3D imaging configuration. The image slices of the involved region in the modified image in DICOM format are preprocessed first using developed a Matlab code, which is an open source medical software used to reconstruct structures of the human body based on three-dimensional images which are acquired using CT or MRI images. The proposed algorithm FBIAMV (Fusion Based Integrated Advance Magnetic Visualization) of MRI 3D images generates the three-dimensional models equivalent to different parts of the human body. The proposed multiform method help doctors and other clinicians in diagnosis of diseases leading to a better treatment.
Image fusion process combines the fusion of two or more images into a single image and gives a production image that have more information than any of the input images. In the medical field, it is performed to examine and make better judgments about a person's ailments. Image fusion integrates the (King, 2007) the multi sensor, multi temporal and multitier in order to ensure that the image contains more information.
Advanced method of image processing and analysis the come across using various methods in medicine. In medical applications, image data are used to collect particular information from the patient’s image to know whether it is a curable or non curable disease process or a normal physiological process. Therefore it is obvious that medical images are very significant in today's health care. The images made in medical applications are many-sided and vary differently on request by the applications. 3D medical images show characteristic in a series about the physiological possessions of the structures of the organs. One of which has high spatial resolution and the other has high spectral resolution.
In order to have high resolution medical images for reliable diagnosis, the indulgence of the image is necessary. The scope of image indulgence and test is useful for medical applications to get improvement in the obtained images and to take out quantitative steps in arranging to form a medical image data in a wellorganized and precise way.
In 3D view Medical image development, high level medical equipments were placed into use in the medical field. Enhanced medical images are favored by a surgeon to help analysis and its understanding, since medical image quality are often deteriorated by noise and other data absorbing devices, light situation, etc. Also the target of medical image development is more important for low dissimilarity and high level noise in a medical image. Medical image development technologies have concerned a lot of studies (Rudd et al., 2007). The basic color space (RGB) change to other color spaces is important for mostly edges improvement of the medical color image (El Naqa, 2008; Yoo, et al., 2002).
Pre-processing of MRI images is the main step in image psychoanalysis, which is used for plummeting image noise, highlighting edges, or display digital images. Many noise reduction techniques are used to improve the image quality and, then some morphological operations are practical to notice the cancer cells in the image. KSL Filtering, Median Filter, High Pass Filter, Adaptive Filter, Mean Filter, etc., techniques are used in preprocessing to reduce noise and image enhancement. The enhancement stage includes resolution enhancement and contrast enhancement. These are used to hold back the noise and imaging of false parameters. After this stage the medical image is rehabilitated into standard image without noise.
Thirdly, noise in the occurrence field was shortened by the soft-threshold method. Fourthly, high-frequency coefficients were improved by dissimilar weight principles in dissimilar sub-images (Smith et al., 2007; Bradley et al., 2004). Then, the improved image is obtained through the inverse wavelet change and opposite HAAR transform. Lastly, the filters are applied to sharpen the image, the resulting image is then subtracted from the original image. Experiments show that this technique can not only improve an image's details, but can also protect its edge features efficiently (Pai et al., 2015) .
Yang et al. (2017) proposed a non-all-encompassing analytic radiology and image-guided radiotherapy has witnessed growing attention in applying dissimilar imaging modalities to stage and limit many-sided diseases, such as atherosclerosis or cancer. It has been experiential that using complementary in order from multimodality images often significantly improves the heftiness and rightness of target volume meaning in radiotherapy action of cancer. In this work, a method and an interactive software tool to support this new framework for 3D multimodality checkup image segmentation was presented (Milker-Zabel, 2006). Abood (2013) proposed a study to create the obvious the limits of color medical images by increasing the thickness to get rid of soft edges and some areas that do not approach into view when the edge is selected (El Naqa, 2008; Townsend, 2008).
Recently, the Mobile Access points (MAs) a cross the system is to be collected directly from individual sensors. While simplifying the routing procedure, a major curb with SENMA is that data broadcast is limited by the physical speed of the MAs and their route length, resulting in low throughput, and large delay (Smith et al., 2007). Toloza et al. (2003) proposed a method which gives easy teaching to get better the Medical imagery by MATLAB. Medical images are one of the basic images, since they are used in more receptive field which is a medical field. The main goal is to get better skin and gain better independence of medical images for a right analysis. The future technique by using middle filter for removing noise on the images is followed by a sharp mask filter, which is type of sharpening (Toloza et al., 2003; Smith et al., 2007).
Program runs slowly due to slower performance in 2D analysis, e.g. no purpose for in order pre-processing provide a semiautomatic image segmentation feature. This segmentation is completed by Hounsfield scale, a quantitative scale that describes radio density. The scale sets grayscale principles according to the width of the region to be segmented, such as air, fat, water, muscle contrast and bone (Nestle, et al., 2005; Hu et al., 2001). Many kinds of imperfection may be set up on the image before segmentation. Some are due to the noise generated through the buzzing material (amalgam, titanium plates, shrapnel from firearm bullets, etc.) set up in the patient's body (Jain et al., 1999; Bezdek, 1981). In other cases, tomography gear without proper calibration make images in which firm tissue and soft tissue density have the same grayscale values, i.e., low contrast. For example; imperfectly segment some piece of soft tissues when it should select only hard tissue (bone) (Bezdesk, 1981; Jain et al., 1999).
Image fusion Entropy is a quantitative gauge which is introduced by Shannon to quantify the image. Entropy is differentiated as the quantity of the restricted signal in the concept of EN that has been working in many scientific fields as well as in image processing method and it contains the in order content of an image. Entropy is a limit to evaluate the information amount limited in an image.
The researcher's ultimate aim is to view the MRI image in three Dimensional view with different parameters using recent medical image transformation technique that creates 3D images from sets of 2D slice, which can be determined by a diversity of modalities, such as CT, MRI, ultrasound, etc. Each type of scanner has its own independence due to considerable main attitude of image recording. The general code of 3D reconstruction has the following steps.
Step 1: 2D data slices require being positioned and set exactly with the genuine spatial position, resulting in a data volume. This data volume is saved in other of computers.
Step 2: Use depiction technique to imagine data volume as 3D image. Usual depiction technique for DICOM image are Advanced Multiplanar Rendering (AMPR), Sender Surface Rendering (SSR), and Auto Volume Rendering (AVR) (Milker-Zabel et al., 2006; Pham et al., 2000).
The process of visualization of 3D MRI images is as shown in Figure 1. MRI does not require a great deal calculation, so it is suitable for low pattern computers. This technique can be second-hand to re-slice arrangement, i.e. with axial slices, the MPR technique to re-slice according to unlike instructions, such as coronal, sagittal, or varied SSR method consists a 3D object as a set of surface called Axial surfaces. Each surface contain points which have the same strength on all slices can be used. This method is used to see the surface of an arrangement connectedly from near structure, e.g. skull from slices of head, blood boat scheme from slices of body, etc. SSR method is often second-hand for high dissimilarity data. Two main methods to rebuild surface can be well intended as follows. Contour based rebuilding contours, which are take out from each slice can be connected to make surfaces (Toloza et al., 2003; Abramoff et al., 2004).

Figure 1 . Proposed Methodology Flow Diagram
AVR method is ultra speed technology to view the whole amount of 3D image in rendering Images will be performing by predictive rays from side to side quantity data. Along each ray, the training mechanism and color is required at every image. Then in future, the end to end distance of each ray will to be collected to a pixel on picture plane. This technique helps us to see at distance end to end, a whole dense agreement of the object. One of disadvantages in this technique is vast quantity of calculation, which requires strong prototype computers. This method is appropriate for low contrast data (El Naqa, 2008; Rudd et al., 2007).
The two main methods for IAVM extrapolative can be considered as follows:
Prognostic MRI goes from side to side volume from rear to front from quantity to image plane.
Projecting rays go from side to side quantity from front to back from image plane to volume. There exists some other method to compound image, which is a suitable method depending on the user's purposes. Some usual method in medical image are MIP (Maximum Intensity Projection), Min IP (Minimum Intensity Projection), AC (Alpha Compositing), and NPVR (Non-Photorealistic Volume Rendering) (Cong and Linh, 2009; Yoo et al., 2002; El Naqa et al., 2007).
Advanced tool of MRI Software for reconstructing DICOM image from a set of CT images was built by using Matlab code and Image Processing tool. A DICOM image contains high incidence noise that cannot be reproduced without an error by using any medical replica software. Thus it is essential to filter/reconstruct the images, firstly although on Matlab code and then additional procedure using Invesalius open source medical model software (Sherekar and Pawar, 2014; Jain et al., 1999). An algorithm is a technique for complexity solving using a finite series of orders. In computer science, algorithms process information using numerical operation (El Naqa et al., 2007). In radiology, the algorithms’ processing order on behalf of tissue and radiological width of conservative MRI and CT and on above all make images by mapping quantitative penalty as a gray-scale or color limit. The configuration of new and modified visual image by applies mathematical algorithms using MATLAB to the unique data is called a reconstruction technique. Many techniques have been developed to post process CT volumetric data (Hsu et al., 2007). The simplest technique extracts one single limit of the volumetric data and creates two dimensional (2D) reconstructions in the desired structure. The most usually used simple methods are: the standard out crop, MIP, and Min IP. Higher process volumetric data, and creating a complicated 3D model are additional features for visualizing complex structures (Bezdek et al., 1981; Pham et al., 2000).
Advance Mat lab generates 3D medical imaging analysis based on a sequence of 3D DICOM files acquired with CT or MRI equipments. The main user interface of the software is functionally similar to some expert medical image softwares. It contains a monitor on the right to show pre-processing using MATLAB programming. Figure 2 illustrates the flowchart arrangement of the software for preparing 3D anatomical model for original and processing images. This diagram can be displayed in dissimilar modes. General Toolbars are located on the top and control panel on the left (Townsend, 2008).
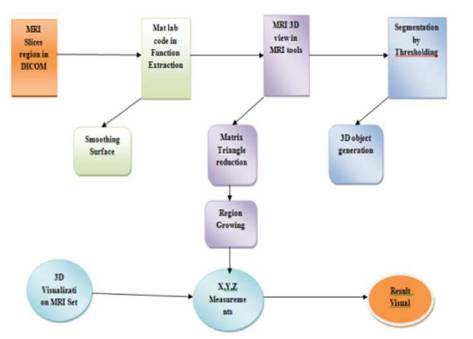
Figure 2. Proposed Algorithm FBIAMV Working Mechanism
Thresholding is one of the majority basic image segmentation methods. The proposed method is commonly used for delineating objects with high contrast with respect to the nearby image background. Threshold standards might be chosen experimentally. For example, to detect MRI 3Ds in PET based on cutoff of Standardized Uptake Value (SUV) (Zheng et al., 2005), a threshold value is more often than not selected as SUV > 2.5 or 40% of maximum SUV. In another example, an optimal threshold representation strength value was chosen iteratively to divide the lungs from the body and chest wall structures. In this work, the thresholding technique to hold up multiple images in the method holds different threshold principles that might be practical to the dissimilar images and the results for different images are joint in rational ways to form the final extended result (El Naqa et al., 2007; Townsend, 2008). The thresholding circumstances idea to explain such manifold image operation were used. For instance, could be “Im1 < 100 and Im2 > 50 | 20 < Im3<150”, where Im1, Im2, and Im3 denote the strength principles of images 1, 2, and 3, respectively. This software tool is able to appreciate the intelligence of such a thresholding state, which performs look and carry out an all the arithmetical and logical calculations to yield the last combined result. In fact, the software tool is implemented with MATLAB and it accepts any valid MATLAB expression as a condition.

Function has to perform the four subtasks as follows (Kleut et al., 2006).
Magnetic Resonance Imaging (MRI) is the major method to diagnose the brain MRI 3Ds and to monitor their treatment. Dissimilar MRI modalities of every method are acquired and these images are interpreted by computerbased image examination methods in order to switch the complexity as well as restraint on time and objectiveness. In this work, the two major novel approaches for analyzing MRI 3D-bearing brain images in a routine way are available. Multi-modal tissue classification with included regularization can segment healthy and pathologic brain tissues counting their sub-compartments to give quantitative volumetric (BauEr, 2013). The method has been evaluated with good consequences on a large figure of clinical and artificial images. The fast run-time of the algorithm allows for an easy adding into the clinical workflow. An additional features has been future for built-in segmentation of longitudinal patient studies, which has been a charge on a small dataset from a multi-center clinical trial with talented outcome.
Fusion Based Integrated Advance Magnetic Visualization of MRI 3D images Algorithm.

The Result is tumor or non tumor segmented a particular boundary image.
The algorithm that was proposed is as follows:

Let X1 … XM are N data points in the input image, let k be th number of Region which is given by the user. Choose R1 …. RK as Region of centers. Distance between each pixel and each cluster centre is found. The distance function is given by,
J=| Xi - Rj | for i=1,…, N and for j=1,…,k, where |Xk –Rk |, the absolute difference of the detachment among a information tip and the cluster centre indicates the distance of the N data points from their respective cluster centers.
Distribute the data points x among the k clusters using the relation,
X ∈ Ri if |x-Rj | < |X-Rk | for i=1,2 ….. R where X denotes the set of data points with Region cluster centre.
Following segmentation and discovery of the preferred area, there are chances for missing clustered regions to occur after the segmentation algorithm, hence morphological filtering is performed for enhancement of the tumor detected portion. Here structuring element used is disk shaped (Hu et al., 2001).
Validations Histogram processing (El Naqa, 2008) is the foundation for much spatial area dispensation technique. Histogram manipulation can be efficient for image enhancement. The straight axes of each histogram plot matches to gray level values. The perpendicular axis correspond to principles of h (rk) = nk or p(rk) = nk / n if the values are normalized. In a dark image, the mechanism of the histogram is intense on the low side of the gray scale. Similarly, the mechanisms of the histogram of the bright picture are biased in the direction of the high side of the gray scale. An image with low difference has a histogram that will be thin and will be centered in the direction of the middle of the gray scale. Finally, the mechanism of the histogram in the highcontrast image wraps a wide range of the gray scale and, in addition to that shares pixels that are not too far (Smith et al., 2007; Milker-Zabel et al., 2006).
The program is evaluated by these criteria (Lin and Lih, 2007).
Segmentation of MRI-bearing brain images is a demanding task for more than a few reasons. Firstly, highgrade 3D usually shows indistinct and unequal limits with discontinuities. Difference uptake and image gaining time after difference inoculation can vary, which change unwanted cells look doubtful if and how the non-image able part of the MRI is imaginary to be handling by segmentation algorithms. As well, MRI 3D sub regions can only be separated when several modalities are combined, which requires precise register as a preprocessing step. Finally, clinical datasets are more often than not acquired in an extremely anisotropic way, most important to a much higher intra-slice order than interslice resolution. These cause problems not only with partial-volume for segmentation, but also registration and re-sampling of the dissimilar modalities in an ordinary space of position, which illustrates the major block used throughout the segmentation pipeline of most algorithms.
Deformable model makes use of restricted independence or edge detection in the images. In the preponderance cases, a level-set is developed towards the MRI 3D border by penetrating for the main predispose in the image or by employing area property that make use of content-based power and texture prototype to develop an energetic curve towards the MRI 3D border in different MRI modalities. Resulting MRI 3D possibility map from the deviation image flank by T1 (Abramoff et al., 2004; Papademetris, et al., 2006) and T1 which shaped the base for the growth of a region-competition level-set algorithm practically.
The problem of lasting list is a great deal and it is more hard when considering a surgery, since then, non-linear data to be built-in and in most cases it is not sufficient to be relevant for normal register algorithms. One request area for this is to register the pre-operative images to intraoperative images or post-resection MRI images. In order to tackle the difficulty of non-rigidly register intrafunctioning MR images are to be sent for pre-operative scans. They are able to grip the automatic brain bond during surgery with a patient constituent technique (FEM) and a non-rigid block-matching technique. Strict constraint on the calculation time might be handled by a parallelized completion and pre-computation of a big part of the dispensation channel. They extra preoperatively acquire MRI modalities, so that in order could be exploited throughout surgery. They achieve near realtime computational pace and were able to give improved vision throughout surgery. A patient biomechanical replica of the brain bend is obtainable and it requests to image register. In their technique, they create and make use of patient non-linear bio-mechanical property for integrating FEM analysis into non-rigid register of pre-operative and intra-operative images. Non-rigid register is relied on to allowing for change after a brain MRI 3D treatment. They formulate it as a maximum a posteriori (MAP) problem and solve it in an EM structure. This way, the likelihood term of the alteration could be seen as a resemblance metric, for which a pointer map could model an assumption for tissue classes (Bradley et al., 2004; Milker-Zabel et al., 2006).
A set of appropriate algorithms were obtained and a new software tool is used for MRI 3D analyzer segmentation of the descriptions from dissimilar imaging modalities. The fundamental code in this work is to unite balancing in order from dissimilar imaging source for improved kind of the work of the imaged obsession pertinent to the depicted clinical task. The multiple images to be built-in into such a framework require not being from dissimilar image modalities.
They might be from the same image modality, but with unlike acquisition protocols (e.g., different MRI pulse sequences), or from the same ahead protocol, but at dissimilar times (e.g. 3D-MRI images). In many situations, one segmentation technique may not perform very well. Different segmentation algorithms may do better for dissimilar issue types, different image gradient situation, different imaging modalities, or even different regions of the same image (King, 2007; El Naqa, 2008).
This plan is embedded in the MRI software tool. There are many dissimilar segmentation algorithms like FBIAMV – Fusion Based Integrated Advance Magnetic Visualization of MRI 3D that has advantages and disadvantages. There are surely other automatic algorithms that could be completed to hold up such a manifold image segmentation framework. The MRI software tool is intended to show these complete algorithms, and to hold up and test the idea of combined algorithms for is similar segmentation methods in to the end to attain better segmentation results. GUI for simulation using 3D toolbox is shown in Figure 3.
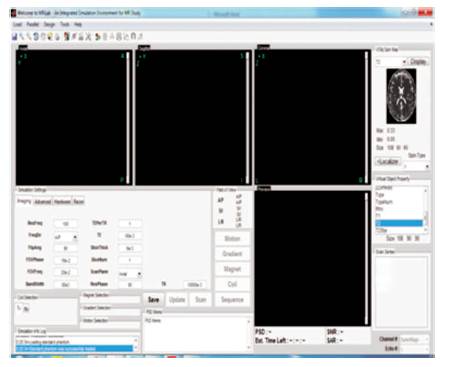
Figure 3. Simulation GUI using MRI 3D Tool Box
The overview of a normal brain is import as a DICOM image and a lots of additional image files format as well as MATLAB type files were analyzed. There are dissimilarity between 2D and 3D picture arrangement, since not all image file format is a 3D. For 2D, the holdup file format comprise DICOM, IMA, MATLAB, and any other usual image file formats like PNG, JPG, TIFF, etc. For 3D datasets, the software tool use DICOM and MATLAB files. Spatial in order, like pixel spacing and slice width are mechanically read from the DICOM files. Other unconfirmed imagery can be loaded into MATLAB software tool by using a MATLAB built-in function or user distinct purpose for other imaging platforms, such as Interfile or Analyze. In any case, for example if pixel spacing, is not available from the image file, the user will be prompted to enter the in order manually (Zheng et al., 2005; Yu-qian et al., 2006). Figure 4 shows dataset selection.
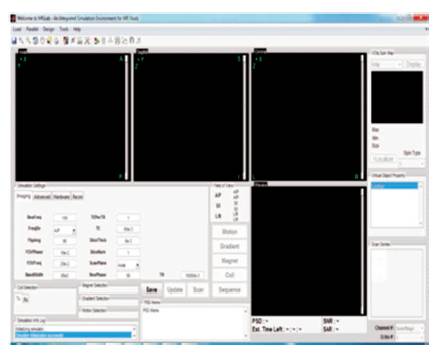
Figure 4. MRI 3D Image Data Set Selection
Images of unlike modalities are frequently acquired from different scanners at different times. Such images are often not only a skew, but could also have dissimilar dimensions and dissimilar image size (number of in each 3D direction). Therefore, image register is an essential step to make sure that the mainfold imagery are correctly allied.
Before the segmentation procedures are practical, images at present hold up some basic routine and physical rigid register method. The first image is defaultly chosen as the orientation picture (the fixed image). All other images are registered unconnectedly to the reference image.
ITK image register algorithms have been implemented and bridged from C and C++ to Matlab. This work has been included into as it presents only built-in method - affine rigid register using joint in order metric, similarity 3D transform and inclined fall optimizer. In this method, joint in order calculation is based on Mattes' method (El Naqa et al., 2007; McAuliffe et al., 2001) .
It also supports change only in the physical list. The panel of directional button is noticeable in the GUI wheel, with respect to in the way of shifting the image to compass reading image. For 3D images, the image shifting way is reliable with the present picture. As explained earlier, all other images are list with respect to the reference image. There is an internal change medium connected with every image. Users can edit any of these alteration matrixes to reach any type of affine alteration. If necessary, the user might also use other available image register tools, to register and then to ensure alteration matrix into MRI. The authors also plan to include multimodality deformable registration algorithms as part of this software in their future work (Yang et al., 2017; McAuliffe et al., 2001; Hsu et al., 2007; Toloza et al., 2003).
Re-sampling, 3D cropping, and 3D zooming are implemented for geometrical processing of the images. The crop characteristic helps to decrease the image measurement so as to save the calculation time. The zooming characteristic helps to set the center of the image to a preferred exaggeration level and to resample the imagery to different images resolutions as shown in Figure 5.
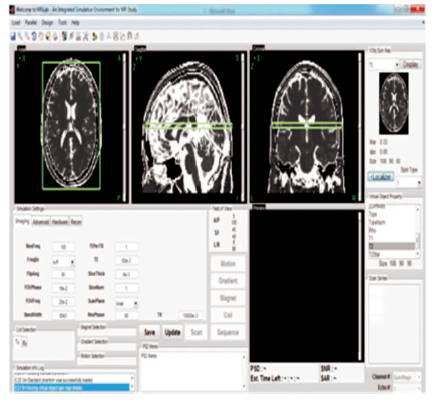
Figure 5. 3D MRI Visualize in Different Angles
Image Intensity preprocessing steps are significant, particularly for image based algorithms. The obtainable preprocessing filter with Virtual Object Property comprise a Gaussian low pass smooth filter, edge preserve smoothing filter, histogram equalization filter, Contrast- Limited Adaptive Histogram Equalization (CLAHE) filter, and a MR image intensity heterogeneity correction filter.
The idea for image segmentation using Virtual Object Property is to sequence the images into dissimilar partition, one by one. The already segmented picture regions is presented from segmentation dispensation. User communication is an important part of the software design. In many cases, users may find that physical interference using physical restriction tools (to be discussed in the next section) may provide help for following auto-segmentation request (Abramoff et al., 2004; Pham, 2001).
Manual Image loading is very significant in any image segmentation software since the previously existing segmentation algorithms may frequently fail to make satisfactory results in many situations. Physical contouring is also used to define the ROI, and to initialize the Active Contour algorithm. Manual loading is used as a knowledge tool for Multimodality segmentation researches to evidence and better appreciate expert's knowledge (Bezdek, 1981; Pham et al., 2000).
MRI tool supports three basic physical contouring methods: continuous manual contouring, line segment manual contouring, and manual imaging. Manually image contour could be additionally considered (Bradley, 1981). For 3D image, physical contours on a discontinuous portion might be interpolated to make the contour for the portions in between.
Localizer can exhibit two to three images at the same time in dissimilar color channels (Yu-qian et al., 2006). At first, the overlay images are intented for register evaluation, where this characteristic is helpful in manual contouring since it allows the user to see more than one image at the same time and to improve the differentiated structure limits from multiple images.
When an automatic segmentation algorithm is used at a step, the consequences of the algorithm will be displayed as a provisional contour. Users can also corroborate the result or discard it. In such a way, users have the alternative to try dissimilar algorithms or dissimilar parameters of the same algorithms (Yoo et al., 2002; Papademetris et al., 2006). Figure 6 shows MRI images in different variations by changing in different angles.
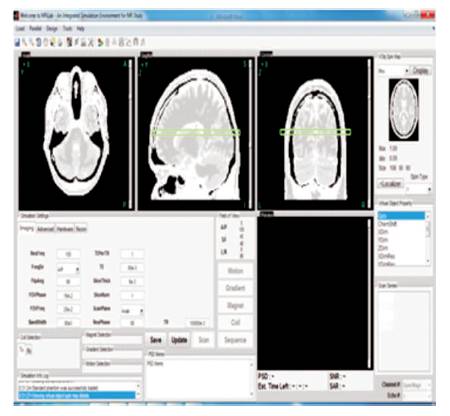
Figure 6. 3D MRI Image Different Axis Variations
The MRI tool has many other useful features to facilitate better image segmentation tasks. The followings are a few important ones (Yang et al., 2017).
Users can surely describe and redefine ROI using the provided physical contouring tools and the curve interpolation feature. The software tool uses the ROI in order to speed up the calculation for the automatic segmentation algorithms by restricting the computation within the ROI region.
Users can put aside the total working status, count all images, settings and current segmentation results, into a project file. The saved project file can then be loaded in a later sitting so that the preceding incomplete work can be continued.
Both the previous preprocessing and post-processing step can be undone, for the last step only. This gives the user a second chance if this last step did not give predictable results.
MRI supports simple 3D image viewing. Users can choose the portion of the figure to view, and can view in a superiorinferior, lateral, or anterior-posterior sides. The software can also represent the segmentation results as a 3D object by using the 3D surface method available with Matlab.
Image window height can be changed for different purposes. The window height location can also be used to change the image strength. Segmentation consequences by automatic segmentation algorithms may be better by these difference improved images (El Naqa et al., 2007; Sebbahi et al., 1997).
An improved subject 3D is that a number of custom algorithms use a group of consumer configurable boundary and users cannot with any difficulty make a choice on the best boundary values. The authors have chosen non-payment main beliefs for the parameter base on the information, which is to be limited to first guess by the user. However, in a lot of scenarios this limit might still need to be tweaked by the user to attain best results. Corroboration of the last result is one of the most difficult tasks in medical image segmentation applications. It could be done by contrasting the segmentation results to histological data if such data are available and appropriate. Results can also be contrasted to other “MRI 3D” standards. However, such contrast is often overstated by other issues, for example, inter-observer randomness. However, it is comparatively safe to think about such routine methods as 'second readers' that would aid the users. This has been shown in many Matlab applications that are helpful to this approach. Image misalignments often occur. For this reason, they have considered a rigid register algorithm in MIR (Jain, et al., 1999; Bezdek, 1981). Figure 7 shows the resultant 3D image.
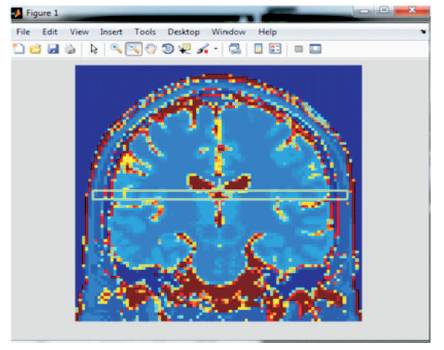
Figure 7. Result in 3D MRI
The research work on modeling medical MRI 3D images, i.e. modified medical implants have been proposed in order to provide the important biomedical image processing and to expand the software tool. The authors have presented a framework to extend dissimilar singlemodality image using FBIAMV – Fusion Based Integrated Advance Magnetic Visualization of MRI 3D images using algorithms to hold up concurrent multi modality representation tasks. In addition, they have designed and developed an interactive software tool “AMRI” to demonstrate the proposed multi modality framework. The AMRI tool allows the user to be relevant to the different segmentation methods at different steps so that advantages of the unusual methods could be compiled and the overall segmentation accuracy could be improved. AMRI is the future of an open source software tool to promote and advance investigations in the medical multimodality imaging analysis and new codes for reconstructing 3D images from a set of MRI images have been developed.